compute gyration/molecule command¶
Syntax¶
compute ID group-ID gyration/molecule general_keyword general_values keyword value ...
- ID, group-ID are documented in compute command
- gyration/molecule = style name of this compute command
- general_keywords general_values are documented in compute
- zero or more keyword/value pairs may be appended
- keyword = tensor
tensor value = none
Examples¶
compute 1 molecule gyration/molecule
compute 2 molecule gyration/molecule tensor
Description¶
Define a computation that calculates the radius of gyration Rg of individual molecules. The calculation includes all effects due to atoms passing thru periodic boundaries.
Rg is a measure of the size of a molecule, and is computed by this formula
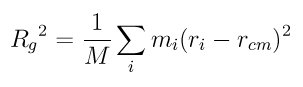
where M is the total mass of the molecule, Rcm is the center-of-mass position of the molecule, and the sum is over all atoms in the molecule and in the group.
If the tensor keyword is specified, then the scalar Rg value is not calculated, but an Rg tensor is instead calculated for each molecule. The formula for the components of the tensor is the same as the above formula, except that (Ri - Rcm)^2 is replaced by (Rix - Rcmx) * (Riy - Rcmy) for the xy component, etc. The 6 components of the tensor are ordered xx, yy, zz, xy, xz, yz.
Rg for a particular molecule is only computed if one or more of its atoms are in the specified group. Normally all atoms in the molecule should be in the group, however this is not required. LIGGGHTS(R)-PUBLIC will warn you if this is not the case. Only atoms in the group contribute to the Rg calculation for the molecule.
The ordering of per-molecule quantities produced by this compute is consistent with the ordering produced by other compute commands that generate per-molecule datums. Conceptually, them molecule IDs will be in ascending order for any molecule with one or more of its atoms in the specified group.
Warning
The coordinates of an atom contribute to Rg in “unwrapped” form, by using the image flags associated with each atom. See the dump custom command for a discussion of “unwrapped” coordinates. See the Atoms section of the read_data command for a discussion of image flags and how they are set for each atom. You can reset the image flags (e.g. to 0) before invoking this compute by using the set image command.
Output info¶
This compute calculates a global vector if the tensor keyword is not specified and a global array if it is. The length of the vector or number of rows in the array is the number of molecules. If the tensor keyword is specified, the global array has 6 columns. The vector or array can be accessed by any command that uses global values from a compute as input. See this section for an overview of LIGGGHTS(R)-PUBLIC output options.
All the vector or array values calculated by this compute are “intensive”. The vector or array values will be in distance units.