compute cna/atom command¶
Syntax¶
compute ID group-ID cna/atom general_keyword general_values cutoff
Examples¶
compute 1 all cna/atom 3.08
Description¶
Define a computation that calculates the CNA (Common Neighbor Analysis) pattern for each atom in the group. In solid-state systems the CNA pattern is a useful measure of the local crystal structure around an atom. The CNA methodology is described in (Faken) and (Tsuzuki).
Currently, there are five kinds of CNA patterns LIGGGHTS(R)-PUBLIC recognizes:
- fcc = 1
- hcp = 2
- bcc = 3
- icosohedral = 4
- unknown = 5
The value of the CNA pattern will be 0 for atoms not in the specified compute group. Note that normally a CNA calculation should only be performed on mono-component systems.
The CNA calculation can be sensitive to the specified cutoff value. You should insure the appropriate nearest neighbors of an atom are found within the cutoff distance for the presumed crystal strucure. E.g. 12 nearest neighbor for perfect FCC and HCP crystals, 14 nearest neighbors for perfect BCC crystals. These formulas can be used to obtain a good cutoff distance:
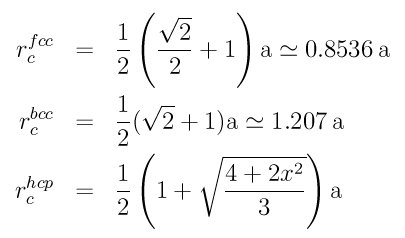
where a is the lattice constant for the crystal structure concerned and in the HCP case, x = (c/a) / 1.633, where 1.633 is the ideal c/a for HCP crystals.
Also note that since the CNA calculation in LIGGGHTS(R)-PUBLIC uses the neighbors of an owned atom to find the nearest neighbors of a ghost atom, the following relation should also be satisfied:
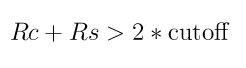
where Rc is the cutoff distance of the potential, Rs is the skin distance as specified by the neighbor command, and cutoff is the argument used with the compute cna/atom command. LIGGGHTS(R)-PUBLIC will issue a warning if this is not the case.
The neighbor list needed to compute this quantity is constructed each time the calculation is performed (e.g. each time a snapshot of atoms is dumped). Thus it can be inefficient to compute/dump this quantity too frequently or to have multiple compute/dump commands, each with a cna/atom style.
Output info¶
This compute calculates a per-atom vector, which can be accessed by any command that uses per-atom values from a compute as input. See Section_howto 15 for an overview of LIGGGHTS(R)-PUBLIC output options.
The per-atom vector values will be a number from 0 to 5, as explained above.
Restrictions¶
none